From genotype to phenotype in complex traits
The DSPR RILs (see the Development of multiparent population resources project page) facilitate systems-level analyses of the genetic architecture of complex traits in several ways: 1) multiple traits can be measured on the same set of RILs, 2) phenotypes can be integrated at multiple different levels, such as gene expression, physiological traits, and visible phenotypes, 3) the genetically identical RILs can be reared in a multitude of different environmental conditions or at different life stages. Our lab has several projects focused using these RILs to learn about different sets of complex traits.
Resource Allocation Phenotypes
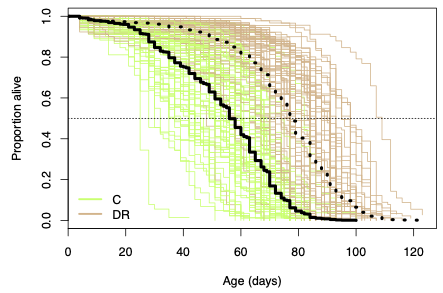
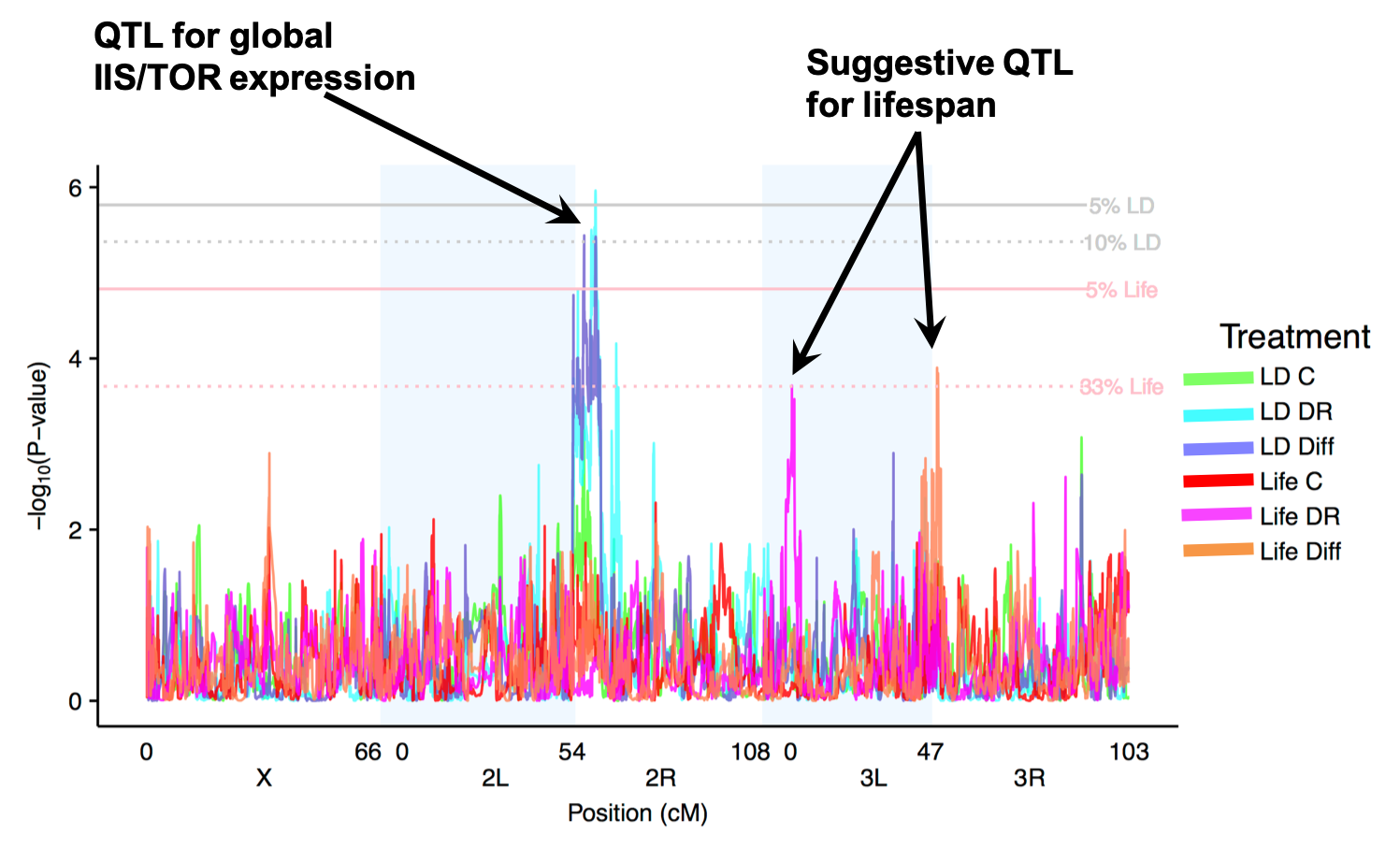
In many ways, allocation strategies are the ultimate complex trait. The allocation of resources is thought to influence nearly all the major structures and functions of an organism, is affected by an array of interacting physiological pathways, varies across the lifetime of the organism, and interacts with many different environmental factors. This underlying complexity makes identifying most of the causative genetic variants very challenging. While the physiological mechanisms underlying the metabolism and allocation of nutrients are fairly well understood and many of the major genes encoding the proteins in these physiological pathways are known, very little is known about the source of natural variation in allocation. We are currently working to measure multiple interrelated phenotypes across multiple levels from gene expression measures to physiological traits (e.g., lipid content, hormone levels, metabolic rate) to life history traits (e.g., median lifespan, fecundity) in multiple nutritional environments using the DSPR RILs with the long term goal of elucidating the relationship between causative genetic variants, environmental influences, transcriptome level phenotypes, physiological phenotypes, and visible phenotypes.
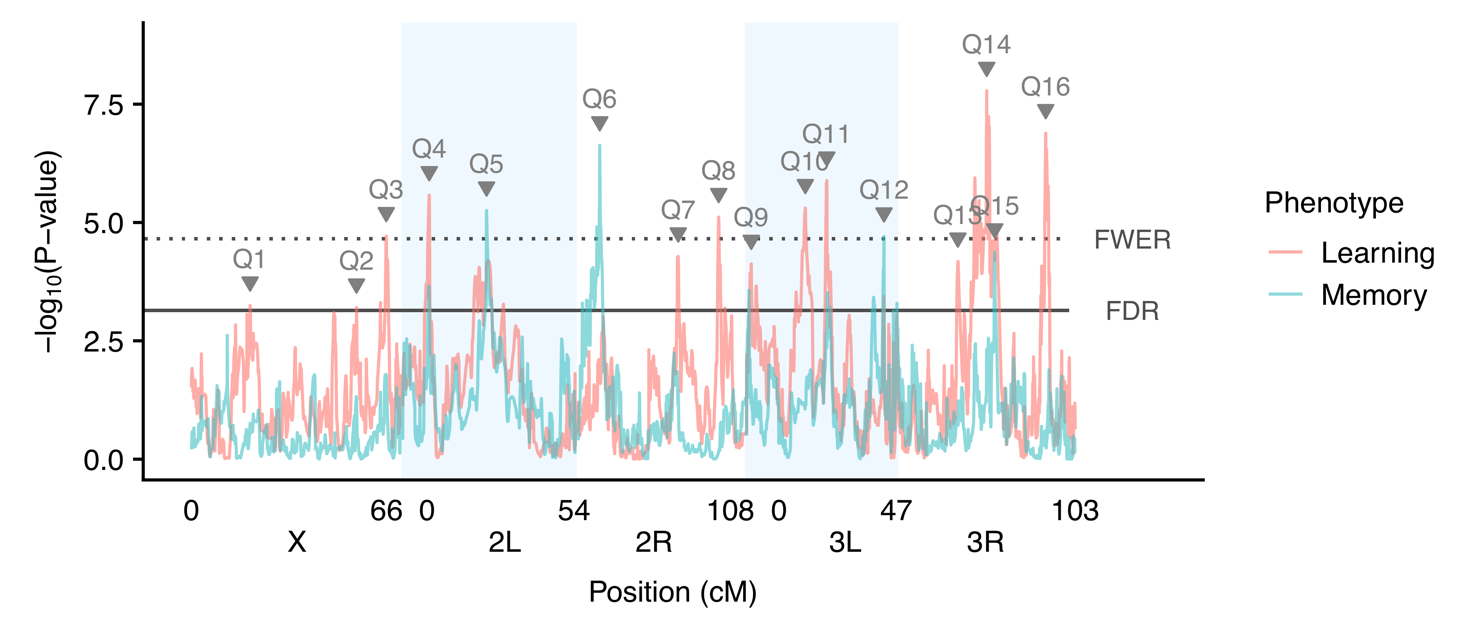
Place Learning and Place Memory
In a second collaborative project with Dr. Troy Zars we are working to identify natural genetic variants influencing place learning and memory. Learning and memory are critical functions for all animals, giving individuals the ability to respond effectively to changes in their environment. Within populations, individuals vary, however the mechanisms underlying this variation in performance are largely unknown. There are very few cases where a segregating genetic variant has been shown to affect any behavior, and none have been identified for place learning or memory. Thus, it remains to be determined what genetic factors cause an individual to have high learning ability, and what factors determine how well an individual will remember what they have learned. Fruit flies can be trained in a “heat box” to learn to remain on one side of a chamber (place learning), and can remember this (place memory) over short timescales. Using this methodology, we have mapped QTL for these traits and are working toward validating the underlying genetic mechanisms at work.